Huilin Li Laboratory
Genome Maintenance and Protein Homeostasis
Living cells rely on large, dynamic molecular machines to replicate DNA, modify and fold proteins, transport proteins to the correct cellular locations, and remove damaged or unnecessary components. Problems with these processes contribute to many human diseases, including cancer, infectious diseases and developmental disorders. Understanding how these molecular systems function at a mechanistic level is essential for identifying new opportunities for therapeutic intervention.
The laboratory of Dr. Huilin Li studies the molecular basis of genome maintenance and protein homeostasis in eukaryotes and pathogens. Major research areas include:
- The mechanisms that ensure accurate DNA replication and coordinate replication with DNA repair
- Enzymatic pathways of protein glycosylation and their roles in protein folding, stability and trafficking
- Chaperone and quality-control systems that remodel or degrade misfolded proteins
By integrating structural, biochemical and cellular approaches, the Li Lab investigates the fundamental principles of cellular organization and regulation, with the goal of translating these insights toward improved treatment strategies.
Our Impact
We're raising thousands to save millions
122
15
10
Huilin Li Laboratory
Huilin Li, Ph.D.
Chair and Professor, Department of Structural Biology; Ralph and Grace Hauenstein Endowed Chair in Structural Biology
Areas of Expertise
Biography
Huilin Li, Ph.D., is an internationally recognized structural biologist with more than 20 years of experience in cryo-electron microscopy (cryo-EM). His current work focuses on the eukaryotic DNA replication, the mycobacterial proteasome system, and the protein folding and protein glycosylation.
Dr. Li earned his Ph.D. in electron crystallography from the University of Science and Technology Beijing, where he trained with the late Prof. K.H. Kuo. He then completed postdoctoral research in the labs of Dr. Bing Jap and Dr. Kenneth Downing at Lawrence-Berkeley National Laboratory, where he studied membrane channels and microtubule structure by cryo-EM. From there, he joined Brookhaven National Laboratory as an associate biophysicist, rising through the ranks to attain a tenured position. In 2010, he joined Stony Brook University as a professor in the Department of Biochemistry and Cell Biology while also maintaining a summer appointment at Brookhaven. He is now a professor and chair of Van Andel Institute’s Department of Structural Biology.
Huilin Li Laboratory
Eukaryotic DNA Replication
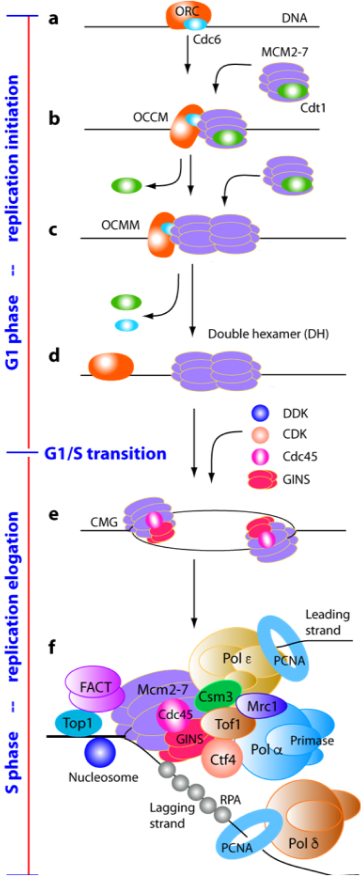
Faithful duplication of the eukaryotic genome requires precise coordination of DNA replication initiation, elongation and termination. Failure to restrict replication to once per cell cycle can lead to genome instability and cancer. Our research uses structural and biochemical approaches to define the molecular mechanisms that ensure accurate DNA replication.
In collaboration with the laboratories of Christian Speck and Bruce Stillman, we elucidated the architecture of the origin recognition complex (ORC) and captured key intermediates in replication origin licensing, including Cdc6-mediated activation of ORC and loading of the Mcm2–7 helicase (Chen et al., 2008, PNAS; Sun et al., 2013, Nat Struct Mol Biol; Sun et al., 2014, Genes Dev; Yuan et al., 2017, Nat Struct Mol Biol; Feng et al., 2021, Nat Commun). We further revealed how two Mcm2–7 hexamers assemble into the pre-replication double hexamer at origin sites (Noguchi et al., 2017, PNAS).
During S phase, the CMG helicase drives DNA unwinding and coordinates with DNA polymerases. In collaboration with Michael O’Donnell’s laboratory, we solved the first structures of the CMG helicase and defined its mechanism of fork unwinding (Yuan et al., 2016, Nat Struct Mol Biol; Yuan et al., 2020, Nat Commun). We also determined the holoenzyme structures of the leading- and lagging-strand polymerases Pol ε and Pol δ, as well as the Pol α–primase complex, and showed how the Ctf4 trimer organizes two CMG helicases and Pol α into a replication factory core (Yuan et al., 2019, eLife; Zheng et al., 2020, PNAS; Yuan et al., 2020, 2023, Nat Commun).
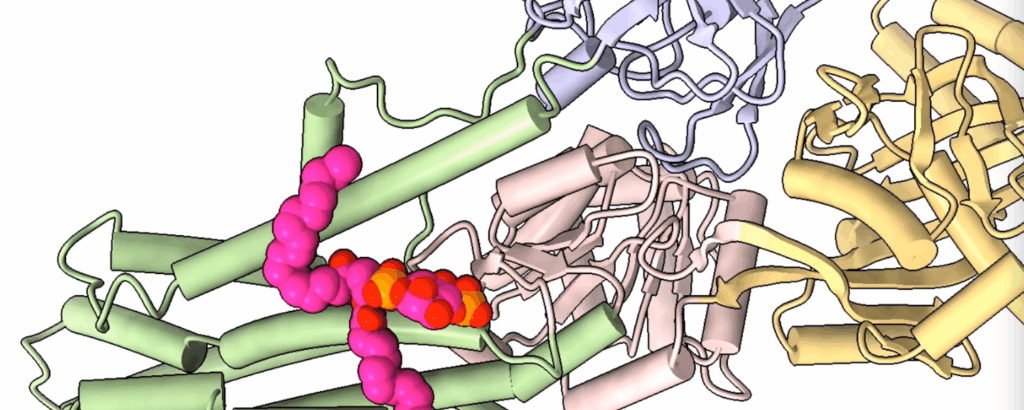
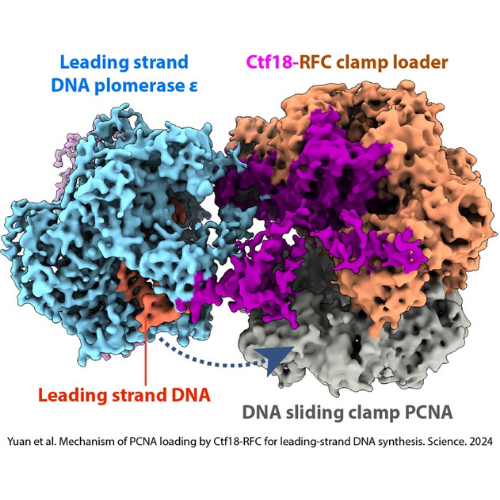
Efficient DNA synthesis requires the PCNA sliding clamp and its loaders. We discovered how the canonical RFC recognizes both 3′ and 5′ DNA ends to load PCNA at gaps during lagging-strand synthesis and DNA repair (Zheng et al., 2022, eLife). We further uncovered how Ctf18-RFC mediates PCNA loading onto the leading strand through direct interactions with Pol ε, enabling lateral DNA transfer into the PCNA ring (Yuan et al., 2024, Science; He et al., 2024, PNAS). Additionally, we revealed how Elg1/ATAD5-RFC acts as a dedicated PCNA unloader through specialized structural features that prevent DNA binding (Zheng et al., 2024, Sci Adv; Wang et al., 2024, Nat Struct Mol Biol).
Finally, we defined the mechanism by which the Rad24-RFC clamp loader installs the 9-1-1 checkpoint clamp onto 5′-recessed DNA, explaining its substrate specificity and role in DNA damage signaling and gap repair (Zheng et al., 2022, Nat Struct Mol Biol; Zheng et al., 2023, Cell Rep).
Together, these studies provide a comprehensive structural framework for eukaryotic DNA replication and its coordination with genome maintenance pathways (Li et al., 2018, BioEssays; O’Donnell et al., 2018, Nat Struct Mol Biol; Yuan et al., 2020, Biochem J).
Related publications
Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H. 2008. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 105(30):10326–10331.
Feng X, Noguchi Y, Barbon M, Stillman B, Speck C, Li H. 2021. The structure of ORC-Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6. Nat Commun 12(1):3883.
Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H*, O’Donnell ME*. 2017. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci U S A 114(5):E697–E706.
*Co-corresponding authors
He Q, Wang F, O’Donnell ME, Li H. 2024. Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC. Proc Nat Acad Sci U S A 121(18):e2319727121.
He Q, Wang F, Yao NY, O’Donnell ME*, Li H*. 2024. Structures of the human leading strand Polε–PCNA holoenzyme. Nat Commun 15(7847).
*Co-corresponding authors
Li H, O’Donnell ME. 2018. The eukaryotic CMG helicase at the replication fork: Emerging architecture reveals an unexpected mechanism. BioEssays 40(3).
Noguchi Y, Yuan Z, Bai L, Schneider S, Zhao G, Stillman B, Speck C, Li H. 2017. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc Natl Acad Sci U S A 114(45):E9529–E9538.
O’Donnell ME, Li H. 2018. The ring-shaped hexameric helicases that function at DNA replication forks. Nat Struct Mol Biol 25(2):122–130.
Sun J, Evrin C, Samel S, Fernadez-Cid A, Riera A, Kawakami H, Zech J, Stillman B, Speck C, Li H. 2013. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol 20(8):944–951.
Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H. 2014. Structural and mechanistic insights into licensing of DNA replication. Genes Dev 28:2291–2303.
Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H*, O’Donnell ME*. 2015. The architecture of a eukaryotic replisome. Nat Struct Mol Biol 22(12):976–982.
*Co-corresponding authors
Wang F, He Q, Yao NY, O’Donnell ME, Li H. 2024. The human ATAD5 has evolved unique structural elements to function exclusively as a PCNA unloader. Nat Struct Mol Biol.
Yuan Z, Bai L, Sun J, Georgescu R, O’Donnell ME, Li H. 2016. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol 23(3):217–224.
Yuan Z, Georgescu R, Bai L, Zhang D, Li H, O’Donnell ME. 2020. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat Commun 11(1):688.
Yuan Z, Georgescu R, Yao NY, Yurieva O, O’Donnell ME, Li H. 2024. Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis. Science 385(6708).
Yuan Z, Georgescu R, Li H*, O’Donnell ME*. 2023. Molecular choreography of primer synthesis by the eukaryotic Pol α-primase. Nat Commun 14(1):3697.
*Co-corresponding authors
Yuan Z, Georgescu R, Santos RLA, Zhang D, Bai L, Yao NY, Zhao G, O’Donnell ME, Li H. 2019. Ctf4 organizes sister replisomes and Pol α into a replication factory. eLife e47405.
Yuan Z, Georgescu R, Schauer GD, O’Donnell ME, Li H. 2020. Structure of the polymerase ε holoenzyme and atomic model of the leading strand replisome. Nat Commun11(1):3156.
Yuan Z, Li H. 2020. Molecular mechanisms of eukaryotic origin initiation, replication fork progression, and chromatin maintenance. Biochem J 477(18):3499–3525.
Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, Speck C, Li H. 2017. Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol 24(3):316–324.
Zheng F, Georgescu RE, Li H, O’Donnell ME. 2020. Structure of eukaryotic DNA polymerase δ bound to the PCNA clamp while encircling DNA. Proc Natl Acad Sci U S A 117(48):30344–30353.
Zheng F, Georgescu R, Yao NY, Li H, O’Donnell ME. 2022. Cryo-EM structures reveal that RFC recognizes both the 3′- and 5′-DNA ends to load PCNA onto gaps for DNA repair. eLife 11:e77469.
Zheng F, Yao NY, Georgescu RE, Li H*, O’Donnell ME*. 2024. Structure of the PCNA unloader Elg1-RFC. Sci Adv 10(9).
*Co-corresponding authors
Zheng F, Georgescu RE, Yao NY, O’Donnell ME, Li H. 2023. Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification on biology. Cell Rep 42(7):112694.
Protein Glycosylation and Trafficking
Protein glycosylation is essential for cell-cell communication and protein folding, stability and trafficking. Our research focuses on the structural and mechanistic basis of protein glycosylation and its coordination with membrane protein biogenesis and intracellular transport.
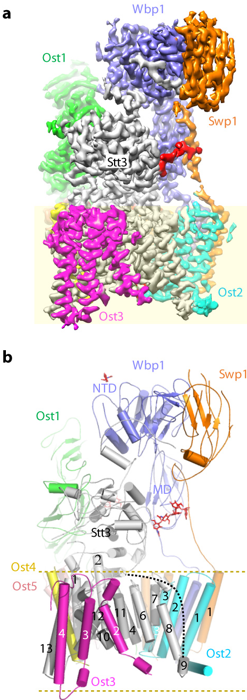
We have made major contributions to understanding N- and O-linked glycosylation in the endoplasmic reticulum (ER). Early work mapped the architecture of the multi-subunit oligosaccharyltransferase (OST) complex and its association with the ribosome (Li et al., 2008, Structure; Harada et al., 2009, PNAS). We later determined the atomic structure of OST, revealing subunit functions and unexpected roles of phospholipids in stabilizing the transmembrane assembly (Bai et al., 2018, Nature). We also showed that protein O-mannosyltransferases share a conserved GT-C fold with OST, establishing an evolutionary link between N- and O-glycosylation (Bai et al., 2019, Nat Struct Mol Biol), and uncovered how Pmt4 selectively O-mannosylates Ser/Thr-rich regions (Du et al., 2025, Nat Commun).
We further investigated O-glucosylation of the Notch receptor, a pathway critical for development and disease. We solved structures of the enzymes responsible for building the O-glucose trisaccharide on Notch, revealing substrate recognition principles (Yu et al., 2016, Nat Chem Biol), resolving a long-standing controversy over the retaining glycosyltransferase mechanism (Yu et al., 2015, Nat Chem Biol), and showing how glycans stabilize EGF repeats (Takeuchi et al., 2017, J Biol Chem). These studies link defects in glycosylation enzymes to cancer and developmental disorders.
In addition, we determined how heparan sulfate polymerization is catalyzed by the EXT1–EXT2 heterodimer, demonstrating that each subunit contributes a distinct glycosyltransferase activity and suggesting a dissociative polymerization mechanism (Li et al., 2023, Nat Chem Biol).
We also study glycan-dependent protein trafficking, including the mannose-6-phosphate (M6P) pathway for lysosomal targeting. Structural studies of the human GlcNAc-1-phosphotransferase revealed a regulatory “hockey stick” motif that controls enzyme activation and explained hyperactivity caused by disease-relevant truncations (Li et al., 2022, Nat Struct Mol Biol; Li et al., 2024, J Biol Chem).
Finally, we investigate how glycosylation is coordinated with membrane protein insertion and trafficking. We determined the structure of the ER membrane complex (EMC), a membrane protein chaperone (Bai et al., 2020, Nature; Bai et al., 2023, Curr Opin Struct Biol), and elucidated lipid flipping mechanisms of multiple P4-ATPase flippases involved in vesicular transport (Bai et al., 2019, 2020, 2021; Duan et al., 2024, J Biol Chem). These studies provide a structural framework for understanding how glycosylated proteins are correctly assembled and delivered within the secretory pathway.
Related Publications
Bai L, Wang T, Zhao G, Kovach A, Li H. 2018. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature 555(7696):328–333.
Bai L, Kovach A, You Q, Hsu C, Zhao G, Li H. 2019. Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p. Nat Commun 10(1):4142.
Bai L, Kovach A, You Q, Kenny A, Li H. 2019. Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex. Nat Struct Mol Biol 26(8):704–711.
Bai L, You Q, Feng X, Kovach A, Li H. 2020. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 584:475–478.
Bai L, You Q, Jain BK, Duan HD, Kovach A, Graham TR, Li H. 2020. Transport mechanism of P4 ATPase phosphatidylcholine flippases. eLife 9:e62163.
Bai L, Jain BK, You Q, Duan HD, Takar M, Graham TR, Li H. 2021. Structural basis of the P4B ATPase lipid flippase activity. Nat Commun 12:5963.
Bai L, Li H. 2023. Structural insights into the membrane chaperones for multi-pass membrane protein biogenesis. Curr Opin Struct Biol 79:102563.
Du M, Yuan Z, Kovach A, Lyu M, Li H. 2025. Pmt4 recognizes two separate acceptor sites to O-mannosylate in the S/T-rich regions of substrate proteins. Nat Commun 16(1):9726.
Duan HD, Li H. 2024. Consensus, controversies, and conundrums of P4-ATPases: the emerging face of eukaryotic lipid flippases. J Biol Chem 300(6):107387.
Duan HD, Jain BK, Li H, Graham TR*, Li H*. 2024. Structural insight into an Arl1-ArfGEF complex involved in Golgi recruitment of a GRIP-domain golgin. Nat Commun 15:1942.
*Co-corresponding authors
Harada Y, Li H, Li H, Lennarz WJ. 2009. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc Natl Acad Sci U S A 106(17):6945–6949.
Li H, Chapla D, Amos RA, Ramiah A, Moremen KW, Li H. 2023. Structural basis for heparan sulfate co-polymerase action by the EXT1–2 complex. Nat Chem Biol.
Li H, Chavan M, Schindelin H, Lennarz WJ, Li H. 2008. Structure of the oligosaccharyl transferase complex at 12 Å resolution. Structure 16(3):432–440.
Li H, Doray B, Jennings BC, Lee W-S, Liu L, Kornfeld S, Li H. 2024. Structure of a truncated human GlcNAc-1-phosphotransferase variant reveals the basis for its hyperactivity. J Biol Chem 300(9):107706.
Li H, Lee WS, Feng X, Bai L, Jennings BC, Liu L, Doray B, Canfield WM, Kornfeld S, Li H. 2022. Structure of the human GlcNAc-1-phosphotransferase αβ subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation. Nat Struct Mol Biol 29(4):348–356.
Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H, Haltiwanger RS. 2017. O-glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. J Biol Chem 292(38):15964–15973.
Yu H, Takeuchi M, LeBarron J, Kantharia J, London E, Bakker H, Haltiwanger RS, Li H*, Takeuchi H*. 2015. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat Chem Biol 11(11):847–854.
*Co-corresponding authors
Yu H, Takeuchi H, Takeuchi M, Liu Q, Kantharia J, Haltiwanger RS, Li H. 2016. Structural analysis of Notch-regulating Rumi reveals basis for pathogenic mutations. Nat Chem Biol 12(9):735–740.
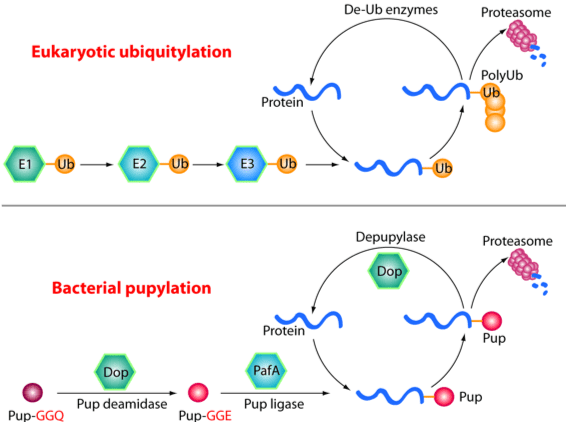
The Pup-proteasome System in Mycobacterium Tuberculosis
Tuberculosis (TB) remains a major global health threat, causing 1.5–2 million deaths each year. Mycobacterium tuberculosis (Mtb), the causative agent of TB, relies on a specialized protein quality control pathway — the Pup–proteasome system — that is functionally analogous but chemically distinct from the eukaryotic ubiquitin–proteasome system. This system is essential for Mtb resistance to host-derived nitric oxide, a key immune defense, making it an attractive target for anti-TB drug development.
Using cryo-EM, X-ray crystallography and biochemical approaches (in collaboration with the laboratories of Carl Nathan, Gang Lin and Heran Darwin), we have defined the molecular architecture and regulation of the Mtb proteasome. We showed that, although structurally similar to the eukaryotic proteasome, the Mtb proteasome employs unique assembly and gating mechanisms (Hu et al., 2006, Mol Microbiol; Li et al., 2010, EMBO J). Proteasomal degradation can be driven by either the ATP-dependent activator Mpa (Wu et al., 2017, Mol Microbiol; Yin et al., 2021, J Biol Chem) or the ATP-independent activator PafE (Jastrab et al., 2015, PNAS; Bai et al., 2016, PNAS; Hu et al., 2018, J Biol Chem). We further discovered that the degradation tag Pup is intrinsically disordered but folds into an α-helix upon binding to Mpa, revealing the basis of substrate recognition (Wang et al., 2010, Nat Struct Mol Biol).
These structural insights enabled structure-guided inhibitor development, leading to the discovery of species-specific inhibitors of the Mtb proteasome, including oxathiazol-2-ones (Lin et al., 2009, Nature), N,C-capped dipeptides (Hsu et al., 2017, Biochemistry), and phenylimidazo- and macrocyclic peptide–based compounds (Zhan et al., 2019, J Med Chem; Zhang et al., 2021, J Med Chem). Notably, some dipeptides selectively inhibit the human immunoproteasome (Santos et al., 2017, Nat Commun).
In parallel, we investigated proteasome-independent protein quality control in Mtb. We defined the disaggregation mechanism of the ClpB hexamer (Yu et al., 2018, PNAS), elucidated how ClpB and DnaK synergistically refold protein aggregates (Yin et al., 2021, Cell Rep) and uncovered an expanded role of GrpE in substrate release from DnaK (Xiao et al., 2024, Nat Commun).
Related Publications
Bai L, Hu K, Wang T, Jastrab JB, Darwin KH, Li H. 2016. Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 113(14):E1983–1992.
Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. 2006. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol 59(5):1417–1428.
Hu K, Jastrab JB, Zhang S, Kovach A, Zhao G, Darwin KH, Li H. 2018. Proteasome substrate capture and gate opening by the accessory factor PafE from Mycobacterium tuberculosis. J Biol Chem 293(13):4713–4723.
Hsu HC, Singh PK, Fan H, Wang R, Sukenick G, Nathan C, Lin G, Li H. 2017. Structural basis for the species-selective binding of N,C-capped dipeptides to the Mycobacterium tuberculosis proteasome. Biochemistry 56(1):324–333.
Jastrab JB, Wang T, Murphy JP, Bai L, Hu K, Merkx R, Huang J, Chatterjee C, Ovaa H, Gygi SP, Li H, Darwin KH. 2015. An adenosine triphosphate-independent proteasome activator contributes to the virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 112(14):E1763–1772.
Li D, Li H, Wang T, Pan H, Lin G, Li H. 2010. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. EMBO J 29(12):2037–2047.
Lin G*, Li D, de Carvalho L, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H*, Nathan C*. 2009. Inhibitors selective for mycobacterial versus human proteasomes. Nature 461(7264):621–626.
*Co-corresponding authors
Santos RLA, Bai L, Singh PK, Murakami N, Fan H, Zhan W, Zhu Y, Jiang X, Zhang K, Assker JP, Nathan CF, Li H, Azzi J, Lin G. 2017. Structure of human immunoproteasome with a reversible and noncompetitive inhibitor that selectively inhibits activated lymphocytes. Nat Commun 8(1):1692.
Samanovic MI, Hsu HC, Jones MB, Jones V, McNeil MR, Becker SH, Jordan AT, Strnad M, Xu C, Jackson M, Li H, Darwin KH. 2018. Cytokinin signaling in Mycobacterium tuberculosis. MBio 19(3).
Wang T, Darwin KH, Li H. 2010. Binding-induced folding of prokaryotic ubiquitin-like protein (Pup) targets substrates for proteasomal degradation. Nat Struct Mol Biol 17(11):1352–1357.
Wu Y, Hu K, Li D, Bai L, Yang S, Jastrab JB, Xiao S, Hu Y, Zhang S, Darwin KH, Wang T, Li H. 2017. Mycobacterium tuberculosis proteasomal ATPase Mpa has a β-grasp domain that hinders docking with the proteasome core protease. Mol Microbiol 105(2):227–241.
Xiao X, Fay A, Santos Molina P, Kovach A, Glickman MS, Li H. 2024. Structure of the M. tuberculosis DnaK−GrpE complex reveals how key DnaK roles are controlled. Nat Comm 15:660.
Yin Y, Kovach A, Hsu HC, Darwin KH, Li H. 2021. The mycobacterial proteasomal ATPase Mpa forms a gapped ring to engage the 20S proteasome. J Biol Chem 100713.
Yin Y, Feng X, Yu H, Fay A, Kovach A, Glickman MS, Li H. 2021. Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system. Cell Rep 35(8):109166.
Yu H, Lupoli TJ, Kovach A, Meng X, Zhao G, Nathan CF, Li H. 2018. ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc Natl Acad Sci U S A 115(41):E9560–E9569.
Zhan W, Hsu HC, Morgan T, Oullette T, Burns-Huang K, Hara R, Wright AG, Imaeda T, Okamoto R, Sato K, Michino M, Ramjee M, Aso K, Meinke P, Foley M, Nathan CR, Li H, Lin G. 2019. Selective phenylimidazole-based inhibitors of the Mycobacterium tuberculosis proteasome. J Med Chem 62(20):9246–9253.
Zhang H, Hsu HC, Kahne SC, Hara R, Zhan W, Jiang X, Burns-Huang K, Ouellette T, Imaeda T, Okamoto R, Kawasaki M, Michino M, Wong TT, Toita A, Yukawa T, Moraca F, Vendome J, Saha P, Sato K, Aso K, Ginn J, Meinke PT, Foley M, Nathan CF, Darwin KH, Li H, Lin G. 2021. Macrocyclic peptides that selectively inhibit the Mycobacterium tuberculosis proteasome. J Med Chem 64(9):6262–6272.
Molecular Mechanisms of Energy Conservation
Modern mitochondria use oxygen as the terminal electron acceptor, but early life evolved in the absence of oxygen and relied on alternative strategies for energy conservation. We study ancestral respiratory systems preserved in the hyperthermophilic archaeon Pyrococcus furiosus, focusing on membrane-bound hydrogenase (MBH) and membrane-bound sulfane sulfur reductase (MBS), which represent evolutionary precursors of modern respiratory Complex I.
We determined the structures of MBH (Yu et al., 2018, Cell) and MBS (Yu et al., 2020, Nat Commun), revealing their redox-coupled ion-pumping mechanisms and evolutionary relationships to Complex I (Yu et al., 2021, JBC review). We also solved the structure of the soluble hydrogenase SHI, a heterotetrameric enzyme in which an NADPH oxidoreductase module (βγ) transfers electrons to a canonical [NiFe] hydrogenase module (αδ) to drive H₂ production (Xiao et al., 2025, Structure), providing a blueprint for future biotechnological applications.
We further investigated flavin-based electron bifurcation (FBEB), a widespread microbial energy conservation strategy that generates low-potential, energy-rich electrons without ATP consumption. In collaboration with the Michael Adams laboratory, we determined the structures of several FBEB-associated complexes, including Fix/EtfABCX (Feng et al., 2021, PNAS), NiFe-HydABCSL (Feng et al., 2022, Sci Adv), and the tungsten-containing WorABSL complex (Feng et al., 2025, PNAS), elucidating their electron transfer pathways.
Finally, we uncovered an unexpected composite flavobicluster bifurcation center — composed of FMN, one [4Fe–4S], and one [2Fe–2S] cluster — in archaeal Nfn-Bfu and bacterial NfnABC enzymes, explaining their ability to catalyze four-electron bifurcation reactions (Xiao et al., 2025, J Biol Chem; Li et al., 2025, Commun Biol).
Related Publications
Feng X, Schut G, Haja D, Adama M, Li H. 2022. Structure and electron transfer pathways of an electron bifurcating NiFe-hydrogenase. Sci Adv 8(8):eabm7546.
Feng X, Schut G, Lipscomb G, Li H, Adams M. 2021. Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX. Proc Natl Acad Sci U S A 118(2).
Feng X, Schut GJ, Putumbaka S, Li H, Adams MWW. 2025. An electron-bifurcating “plug” to a protein nanowire in tungsten-dependent aldehyde detoxification. Proc Natl Acad Sci U S A 122(30):e2501900122.
Li H, Schut GJ, Feng X, Adams MWW, Li H. 2025. Cryo-EM reveals a composite flavobicluster electron bifurcation site in the Bfu family member NfnABC. Commun Biol 14;8(1):239.
Yu H, Wu CH, Schut G, Haja D, Zhao G, Peters J, Adams M, Li H. 2018. Structure of an ancient respiratory system. Cell 173(7):1636-1649.
Yu H, Haja DK, Schut GJ, Wu CH, Meng X, Zhao G, Li H, Adams MWW. 2020. Structure of the respiratory MBS complex reveals iron-sulfur cluster catalyzed sulfane sulfur reduction in ancient life. Nat Commun 11(1):5953.
Yu H, Schut G, Haja D, Adams M, Li H. 2021. Evolution of complex I-like respiratory complexes. J Biol Chem 296: 100740. (Invited review).
Xiao X, Schut GJ, Feng X, McTernan PM, Haja DK, Lanzilotta WN, Adams MWW, Li H. 2025. Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P. furiosus. Structure 33(9):1470-1483.
Xiao X, Schut GJ, Feng X, Nguyen DMN, Huang H, Wang S, Li H, Adams MWW. 2025. Cryo-EM structures define the electron bifurcating flavobicluster and ferredoxin binding site in an archaeal Nfn-Bfu transhydrogenase. J Biol Chem 301(4):108410.
Huilin Li Laboratory
SELECTED PUBLICATIONS
For a full list of Dr. Li’s publications, please visit his NCBI bibliography.
Chen X, Duan HD, Hoy MJ, Koteva K, Spitzer M, Guitor AK, Puumala AK, Fiebig AA, Hu G, Yiu B, Chou S, Bian Z, Choi Y, Guo ABY, Wang W, Sun S, Robbins N, Averette AF, Cook MA, Truant R, MacNeil LT, Brown ED, Kronstad JW, Coombes BK, Cowen LE, Heitman J, Li H, Wright GD. 2025. Butyrolactol A enhances caspofungin efficacy via flippase inhibition in drug-resistant fungi. Cell.
Feng X, Spiering MM, Ruda de Luna Santos A, Benkovic SJ, Li H. 2025. Structural insights into the exchange mechanism of a replicative DNA polymerase. Nucleic Acids Res 53(22).
Du M, Yuan Z, Kovach A, Lyu M, Li H. 2025. Pmt4 recognizes two separate acceptor sites to O-mannosylate in the S/T rich regions of substrate proteins. Nat Commun 16(1).
*Selected as a featured paper
Jain BK, Duan HD, Valentine C, Samiha A, Li H, Graham TR. 2025. P4-ATPases control phosphoinositide membrane asymmetry and neomycin resistance. Nat Cell Biol 27(7):114-1124.
Xiao X, Schut G, Feng X, McTernan PM, Haja DK, Lanzilotta WN, Adams MWW, Li H. 2025. Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P. furiosus. Structure.
Wang F, He Q, O’Donnell ME, Li H. 2025. The proofreading mechanism of the human leading-strand DNA polymerase holoenzyme. Proc Natl Acad Sci U S A 122(22):e2507232122.
Li H, Doray B, Jennings BC, Lee W-S, Liu L, Kornfeld S, Li H. 2024. Structure of a truncated human GlcNAc-1-phosphotransferase variant reveals the basis for its hyperactivity. J Biol Chem 300(9):107706.
He Q, Wang F, Yao NY, O’Donnell ME*, Li H*. 2024. Structures of the human leading strand Polε–PCNA holoenzyme. Nat Commun 15(7847).
*Co-corresponding authors
Yuan Z, Georgescu R, Yao NY, Yurieva O, O’Donnell ME, Li H. 2024. Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis. Science 385(6708).
Feng X, Schut GJ, Adams MWW, Li H. Structures and electron transport paths in the four families of flavin-based electron bifurcation enzymes. In: Harris JR, Marles-Wright J (eds). Macromolecular Protein Complexes V: Structure and Function, Springer-Nature: 383-408.
Duan HD, Li H. 2024. Consensus, controversies, and conundrums of P4-ATPases: the emerging face of eukaryotic lipid flippases. J Biol Chem 300(6):107387.
Wang F, He Q, Yao NY, O’Donnell ME, Li H. 2024. The human ATAD5 has evolved unique structural elements to function exclusively as a PCNA unloader. Nat Struct Mol Biol.
Yuan Z, Li H. 2024. Primase and polymerase α tango to make an RNA-DNA hybrid primer. FEBS J 291(9):1889–1891.
He Q, Wang F, O’Donnell ME, Li H. 2024. Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC. Proc Nat Acad Sci U S A 121(18):e2319727121.
Duan HD, Jain BK, Li H, Graham TR*, Li H*. 2024. Structural insight into an Arl1-ArfGEF complex involved in Golgi recruitment of a GRIP-domain golgin. Nat Commun 15:1942.
*Co-corresponding authors
Zheng F, Yao NY, Georgescu RE, Li H*, O’Donnell ME*. 2024. Structure of the PCNA unloader Elg1-RFC. Sci Adv 10(9).
*Co-corresponding authors
Xiao X, Fay A, Santos Molina P, Kovach A, Glickman MS, Li H. 2024. Structure of the M. tuberculosis DnaK−GrpE complex reveals how key DnaK roles are controlled. Nat Comm 15:660.
Hsu HC, Li D, Zhan W, Ye J, Liu YJ, Leung A, Qin J, Crespo B, Gamo FJ, Zhang H, Cui L, Roth A, Kirkman LA, Li H, Lin G. 2023. Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors. Nat Commun 14(1):8302.
Zheng F, Georgescu RE, Yao NY, O’Donnell ME, Li H. 2023. Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification on biology. Cell Rep 42(7):112694.
Feng X, Spiering MM, de Luna Almeida Santos R, Benkovic SJ, Li H. 2023. Structural basis of the T4 bacteriophage primosome assembly and primer synthesis. Nat Commun 14(1):4396.
Burgie ES, Li H, Gannam ZTK, McLoughlin KE, Vierstra RD*, Li H*. 2023. The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms. Nat Plants 9(7):1116–1129.
*Co-corresponding authors
Hsu HC, Wang J, Kjellgren A, Li H, DeMartino GN. 2023. Ηigh-resolution structure of mammalian PI31-20S proteasome complex reveals mechanism of proteasome inhibition. J Biol Chem 299(7):104862.
Bai L, Li H. 2023. Structural insights into the membrane chaperones for multi-pass membrane protein biogenesis. Curr Opin Struct Biol 79:102563.
Yuan Z, Georgescu R, Li H*, O’Donnell ME*. 2023. Molecular choreography of primer synthesis by the eukaryotic Pol α-primase. Nat Commun 14(1):3697.
*Co-corresponding authors
Wang F, He Q, Zhan W, Yu Z, Finkin-Groner E, Ma X, Lin G, Li H. 2023. Structure of the human UBR5 E3 ubiquitin ligase. Structure.
Li H, Chapla D, Amos RA, Ramiah A, Moremen KW, Li H. 2023. Structural basis for heparan sulfate co-polymerase action by the EXT1–2 complex. Nat Chem Biol.
Bai L, Li H. 2022. Cryo-EM structures of the endoplasmic reticulum membrane complex. FEBS J 289(1):102–112.
Langston LD, Yuan Z, Georgescu R, Li H, O’Donnell ME. 2022. SV40 T-antigen uses a DNA shearing mechanism to initiate origin unwinding. Proc Natl Acad Sci U S A 119(49):e2216240119.
Li H, Lee WS, Feng X, Bai L, Jennings BC, Liu L, Doray B, Canfield WM, Kornfeld S, Li H. 2022. Structure of the human GlcNAc-1-phosphotransferase αβ subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation. Nature Struct Molec Biol 29(4):348–356.
Li H, O’Donnell M, Kelch B. 2022. Unexpected new insights into DNA clamp loaders. Bioessays 44(11):e2200154.
Schut GJ, Haja DK, Feng X, Poole FL, II, Li H, Adams MWW. 2022. An abundant and diverse new family of electron bifurcating enzymes with a non-canonical catalytic mechanism. Front Microbiol 13:946711.
Wang F, Feng X, He Q, Li H, Li H. 2022. The S. Cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly. J Biol Chem 102852.
Xiao X, Feng X, Yoo JH, Kovach A, Darwin KH, Li H. 2022. The β-grasp domain of proteasomal ATPase Mpa makes critical contacts with the Mycobacterium tuberculosis 20S core particle to facilitate degradation. mSphere 7(5):e00227422.
Zheng F, Georgescu R, Yao NY, Li H, O’Donnell ME. 2022. Cryo-EM structures reveal that RFC recognizes both the 3′- and 5′-DNA ends to load PCNA onto gaps for DNA repair. eLife 11:e77469.
Li H*, Burgie S*, Gannam ZTK, Li H#, Vierstra RD#. 2022. Plant phytochromes B is an asymmetric dimers with unique signaling potential. Nature 604(7904): 127-133.
* Co-first authors
# Co-corresponding authors
Li H#, Lee WS#, Feng X, Bai L, Jennings JC, Liu L, Doray B, Canfield WM, Kornfeld S*, Li H*. 2022. Structure of the human GlcNAc-1-phosphotransferase αβ subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation. Nat Struct Mol Biol 29(4): 348-356.
# Co-first authors
* Co-corresponding authors
Zheng F, Georgescu RE, Yao NY, O’Donnell ME*, Li H*. 2022. DNA is loaded through the 911 DNA checkpoint clamp in the opposite direction of the PCNA clamp. Nat Struc Mol Biol 29(4): 378-385.
* Co-corresponding authors
Feng X, Schut G, Haja D, Adama M, Li H. 2022. Structure and electron transfer pathways of an electron bifurcating NiFe-hydrogenase. Sci Adv 8(8): eabm7546.
Hsu HC, Wang M, Kovach A, Darwin AJ, Li H. 2022. Pseudomonas aeruginosa C-terminal processing protease CtpA assembles into a hexameric structure that requires activation by a spiral-shaped lipoprotein-binding partner. mBio 13(1): e0368021.
Zhao P, Zhao C, Chen D, Yun C, Li H*, Bai L*. 2021. Structure and activation mechanism of the hexameric plasma membrane H+-ATPase. Nat Commun 12:6439.
*Co-corresponding authors
Included as an Editor’s Highlight
Bai L, Jain BK, You Q, Duan HD, Takar M, Graham TR, Li H. 2021. Structural basis of the P4B ATPase lipid flippase activity. Nat Commun 12:5963.
Du M, Yuan Z, Werneburg GT, Henderson NS, Chauhan H, Kovach A, Zhao G, Johl J, Li H, Thanassi DG. 2021. Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis. Nat Commun 12(1):5207.
Yu H, Schut GJ, Haja DK, Adams MWW, Li H. 2021. Evolution of complex I-like respiratory complexes. JBC Rev 296:100740.
Yin Y, Feng X, Yu H, Fay A, Kovach A, Glickman MS, Li H. 2021. Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system. Cell Rep 35(8):109166.
Zhang H, Hsu HC, Kahne SC, Hara R, Zhan W, Jiang X, Burns-Huang K, Ouellette T, Imaeda T, Okamoto R, Kawasaki M, Michino M, Wong TT, Toita A, Yukawa T, Moraca F, Vendome J, Saha P, Sato K, Aso K, Ginn J, Meinke PT, Foley M, Nathan CF, Darwin KH, Li H, Lin G. 2021. Macrocyclic peptides that selectively inhibit the Mycobacterium tuberculosis proteasome. J Med Chem 64(9):6262–6272.
Yin Y, Kovach A, Hsu HC, Darwin KH, Li H. 2021. The mycobacterial proteasomal ATPase Mpa forms a gapped ring to engage the 20S proteasome. J Biol Chem 100713.
Bai L, Li H. 2021. Cryo-EM structures of the endoplasmic reticulum membrane complex. FEBS J.
Li H, Zheng F, O’Donnell M. 2021. Water skating: How polymerase sliding clamps move on DNA. FEBS J.
Feng X, Schut GJ, Lipscomb GL, Li H, Adams MWW. 2021. Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX. Proc Natl Acad Sci U S A 118(2).
Bai L, Li H. 2021. Protein N-glycosylation and O-mannosylation are catalyzed by two evolutionarily related GT-C glycosyltransferases. Curr Opin Struct Biol 68:66–73.
Bohl T, Bai L, Li H. 2021. Recent progress in structural studies on the GT-C superfamily of protein glycosyltransferases. Subcell Biochem 96:259–271.
Li H, Yao NY, O’Donnell ME. 2020. Anatomy of a twin DNA replication factory. Biochem Soc Trans 48(6):2769–2778.
Zheng F, Georgescu RE, Li H, O’Donnell ME. 2020. Structure of eukaryotic DNA polymerase δ bound to the PCNA clamp while encircling DNA. Proc Natl Acad Sci U S A 117(48):30344–30353.
Yu H, Haja DK, Schut GJ, Wu CH, Meng X, Zhao G, Li H, Adams MWW. 2020. Structure of the respiratory MBS complex reveals iron-sulfur cluster catalyzed sulfane sulfur reduction in ancient life. Nat Commun 11(1):5953.
Yuan Z, Li H. 2020. Molecular mechanisms of eukaryotic origin initiation, replication fork progression, and chromatin maintenance. Biochem J 477(18):3499–3525.
Yuan Z, Schneider S, Dodd T, Riera A, Bai L, Yan C, Magdalou I, Ivanov I, Stillman B, Li H, Speck C. 2020. Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6. Proc Natl Acad Sci U S A 117(30):17747–17756.
Yuan Z, Georgescu R, Schauer GD, O’Donnell ME, Li H. 2020. Structure of the polymerase ε holoenzyme and atomic model of the leading strand replisome. Nat Commun 11(1):3156.
Yuan Z, Georgescu R, Bai L, Zhang D, Li H, O’Donnell ME. 2020. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat Commun 11(1):688.
Bai L, You Q, Jain BK, Duan HD, Kovach A, Graham TR, Li H. 2020. Transport mechanism of P4 ATPase phosphatidylcholine flippases. eLife.
Bai L, You Q, Feng X, Kovach A, Li H. 2020. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature.
Yuan Z, Georgescu R, Santos RLA, Zhang D, Bai L, Yao NY, Zhao G, O’Donnell ME, Li H. 2019. Ctf4 organizes sister replisomes and Pol α into a replication factory. eLife e47405.
Zhan W, Hsu HC, Morgan T, Oullette T, Burns-Huang K, Hara R, Wright AG, Imaeda T, Okamoto R, Sato K, Michino M, Ramjee M, Aso K, Meinke P, Foley M, Nathan CR, Li H, Lin G. 2019. Selective phenylimidazole-based inhibitors of the Mycobacterium tuberculosis proteasome. J Med Chem.
Bai L, Kovach A, You Q, Hsu C, Zhao G, Li H. 2019. Autoinhibition and activation mechanisms of the eurkaryotic lipid flippase Drs2p-Cdc50p. Nat Commun 10(1):4142.
Li H, O’Donnell ME. 2019. DNA replication from two different worlds. Science 363(6429):814–815.
Bai L, Kovach A, You Q, Kenny A, Li H. 2019. Structure of the eukaryotic protein O-mannosyltransferase Pmt1−Pmt2 complex. Nat Struct Mol Biol.
Bai L, Li H. 2019. Cryo-EM is uncovering the mechanism of eukaryotic protein N-glycosylation. FEBS 286(9):1638–1644.
Hu K, Jordan AT, Zhang S, Dhabaria A, Kovach A, Rangel M, Ueberheide B, Li H, Darwin KH. 2019. Characterization of guided entry of tail-anchored proteins 3 homologues in Mycobacterium tuberculosis. J Bacteriol.
Samanovic MI, Hsu HC, Jones MB, Jones V, McNeil MR, Becker SH, Jordan AT, Strnad M, Xu C, Jackson M, Li H, Darwin KH. 2018. Cytokinin signaling in Mycobacterium tuberculosis. MBio 19(3).
O’Donnell ME, Li H. 2018. The ring-shaped hexameric helicases that function at DNA replication forks. Nat Struct Mol Biol 25(2):122–130.
Du M, Yaun Z, Yu H, Henderson N, Sarowar S, Zhao G, Werneburg GT, Thanassi DG, Li H. 2018. Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD. Nature.
Yu H, Wu CH, Schut GJ, Haja DK, Zhao G, Peters JW, Adams MWW, Li H. 2018. Structure of an ancient respiratory system. Cell.
Li H, O’Donnell ME. 2018. The eukaryotic CMG helicase at the replication fork: Emerging architecture reveals an unexpected mechanism. BioEssays40(3):1700208.
*Highlighted in Advanced Science News.
Bai L, Wang T, Zhao G, Kovach A, Li H. 2018. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature.
Bai L, Yuan Z, Sun J, Georgescu R, O’Donnell ME, Li H. 2017. Architecture of the Saccharomyces cerevisiae replisome. Adv Exp Med Biol 1042:207–228.
Noguchi Y, Yuan Z, Bai L, Schneider S, Zhao G, Stillman B, Speck C, Li H. 2017. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc Natl Acad Sci U S A.
Wu Y, Hu K, Li D, Bai L, Yang S, Jastrab JB, Xiao S, Hu Y, Zhang S, Darwin KH, Wang T, Li H. 2017. Mycobacterium tuberculosis proteasomal ATPase Mpa has a β-grasp domain that hinders docking with the proteasome core protease. Mol Microbiol 105(2):227–241.
Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, Speck C, Li H. 2017. Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol 24(3):316–324.
Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H, O’Donnell ME. 2017. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci U S A 114(5):E697–E706.
Yu H, Takeuchi H, Takeuchi M, Liu Q, Kantharia J, Haltiwanger RS, Li H. 2016. Structural analysis of Notch-regulating Rumi reveals basis for pathogenic mutations. Nat Chem Biol 12:735–740.
Yuan Z, Bai L, Sun J, Georgescu R, O’Donnell ME, Li H. 2016. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol 23(3):217–224.
O’Donnell M, Li H. 2016. The eukaryotic replisome goes under the microscope. Curr Biol 26(6):R247-56.
Bai L, Hu K, Wang T, Jastrab JB, Darwin KH, Li H. 2016. Structural analysis of the dodecameric proteasome activator PafE in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 113(14):E1983-92.
Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O’Donnell ME. 2015. The architecture of a eukaryotic replisome. Nat Struct Mol Biol 22:976–982.
Yu H, Takeuchi M, LeBarron J, Kantharia J, London E, Bakker H, Haltiwanger RS, Li H, Takeuchi H. 2015. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat Chem Biol 11(11):847–854.
Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, Li H. 2014. Structural and mechanistic insights into licensing of DNA replication. Genes Dev 28:2291–303.
Sun J, Evrin C, Samel S, Fernadez-Cid A, Riera A, Kawakami H, Zech J, Stillman B, Speck C, Li H. 2013. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol 20(8):944–951.
Li H, Stillman B. 2012. The origin recognition complex: a biochemical and structural view. Subcell Biochem 62:37–58.
Sun J, Li H. 2010. How to operate a cryo-electron microscope. Methods Enzymol 481:231-49.
Li D, Li Hua, Wang T, Lin G, Li H. 2010. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. EMBO J 29:2037–2047.
Wang T, Darwin KH, Li H. 2010. Binding-induced folding of prokaryotic ubiquitin-like protein (Pup) targets substrates for proteasomal degradation. Nat Struct and Mol Biol 17:1352–1357.
Lin G, Li D, de Carvalho L, Deng H, Tao H, Vogt G, Wu K, Schneider J, Chidawanyika T, Warren JD, Li H, and Nathan C. 2009. Inhibitors selective for mycobacterial versus human proteasomes. Nature 461:621–626.
Huilin Li Laboratory

Biography
Dr. H. Diessel Duan earned his B.S in chemical engineering from Jiangnan University and his Ph.D. in biochemistry from Auburn University. His graduate work focused on catalase-peroxidase (KatG), a key enzyme for bacterial pathogens to survive oxidative damage from host immune responses. He went on to study flavin-based electron bifurcation, now regarded as the third mechanism of biological energy conservation. In 2020, he joined the lab of Dr. Huilin Li as a postdoctoral fellow.

Nancy Duchaine
Senior Administrative Assistant I, Department of Structural Biology

Biography

Qing He, Ph.D.
Postdoctoral Fellow, Li Laboratory
Structural biology of DNA replication and repair
Biography
Dr. Qing He graduated from Shandong Normal University with a B.S. in biotechnology and got her Ph.D. in microbiology from Shandong University. Her graduate work focused on the structural and functional study of c-di-NMP related proteins, and the many biochemical experiments. Dr. He’s main lab competencies lie in cloning, protein purification, crystallization and small molecular and protein interaction tests. She is a postdoctoral fellow in the lab of Dr. Huilin Li at Van Andel Institute.

Hao-Chi Hsu, Ph.D.
Research Scientist, Department of Structural Biology

Sofia Ievleva
Ph.D. Student, VAI Graduate School
Research Focus: Investigating the structural pharmacology of trpm4 channel

Biography
Mentor: Huilin Li, Ph.D.
Research Focus: Investigating the structural pharmacology of trpm4 channel
Experience: Sofia earned her bachelor’s degree in biology from Alcorn State University. She has participated in several research projects, such as exploring curcumin drug treatment of an ovarian cancer cell line, evaluating the economic impacts of childhood obesity, and studying health disparity, increased obesity and cancer-triggering concerns in Mississippi. This latter project was presented at the 4th Annual Advancing Mississippi Conference and detailed in a publication. In addition to her research experience, Sofia has extensive tutoring experience in STEM subjects, supporting students from all academic levels.
Undergraduate Institution: Alcorn State University
Hometown: Voronezh, Russia
Publications
Sarkar S, Taylor A, Dutta P, Bajaj M, Nash J, Ravola M, Ievleva S, Llyod C, Ola P, Jenkins B, Sengupta B, Roy D. 2021. Health disparity and COVID 19: a retrospective analysis. Health Sci Rep 4(3):e345.

Amanda Kovach, B.S.
Lab Manager, Department of Structural Biology

Zhengtao Liu, Ph.D.
Postdoctoral Fellow, Li Laboratory
Meinan Lyu, Ph.D.
Research Scientist, Department of Structural Biology

Chang Sun, Ph.D.
Research Scientist, Department of Structural Biology

Merissa (Xiansha) Xiao, Ph.D.
Research Scientist, Li Laboratory
Proteasome and protein folding system involved in multi-drug resistance in Mycobacteria
Biography
Dr. Merissa Xiao earned her Ph.D. in structural biology and biotechnology from Leiden University and her B.S. in bioengineering from Northeast Agricultural University. Her graduate work focused on antibiotic biosynthesis and drug-target interactions. Dr. Xiao’s main lab competencies lie in molecular biology, chemistry biology and structural biology. In 2021, she joined the lab of Dr. Huilin Li as a postdoctoral fellow.
Qinglong You, Ph.D.
Research Scientist, Department of Structural Biology
Epigenetic mechanisms in DNA damage point
Biography
Dr. Qinglong You earned a B.S. in applied physics from Huazhong University of Science and Technology, and a Ph.D. in molecular biology and Bbiochemistry from the Institute of Biophysics of the Chinese Academy of Sciences. In his graduate work, Dr. You studied nucleosome binding proteins and mBend3 DNA complex with use of X-ray crystallography. Much of his laboratory work includes protein analysis and expression and crystal structure determination. In 2019, he joined the laboratory of Dr. Huilin Li at Van Andel Institute as a postdoctoral fellow.
Fengwei Zheng, Ph.D.
Research Scientist, Department of Structural Biology
DNA replication and repair
Biography
Dr. Fengwei Zheng earned his B.S. in biotechnology from Dalian Ocean University, and his Ph.D. in microbiology from Wuhan University. His graduate work focused on the structural and functional study of CSFV NS3. Along with his other research work, he is skilled in molecular cloning, protein expression and purification, and bioinformatics. In Oct. 2018, he joined the laboratory of Dr. Huilin Li at Van Andel Institute as a postdoctoral fellow.